

Net-CSSP
Neural networks for calculating Contact-dependent Secondary Structure Propensity
Applications in predicting non-native secondary structures and amyloid fibril formation
What is NetCSSP?
NetCSSP is a neural network-based predictor of contact-dependent secondary structure propensity in protein sequences. The local secondary structure of a protein can change depending on the tertiary environment such as inter-molecular interactions and misfolding. One prominent example is non-native beta- strand formation in most amyloid fibrils.
We have quantitatively standardized the tertiary interaction energies of various proteins and then trained neural networks (NetCSSP) to predict secondary structures of a protein from the standardized tertiary interaction energies together with a sequence context (7-residue span) . Thus the power of the Net-CSSP method is that it provides insight on the change of secondary structural propensities in the entire range of tertiary interaction energies for a given amino acid sequence without any knowledge of its 3D structure.
One important application of Net-CSSP is its accurate predictive ability in identifying core sequences of amyloid fibril formation where the switch in local secondary structure occurs.
Dual artificial neural network for CSSP prediction
The dual network architecture consists of the two single networks that use distinct output nodes.
One network has output nodes for a-helix and non-helix, thus predicting helical propensity. The other network has output nodes for b-strand and non-beta strand, thus predicting beta propensity.
The input layer of single and dual networks contains an additional variable node for the query sequence. This variable node was set to the computed energy term (>(i,i±4) energy) of the query sequence for single network architecture. However, for dual network architecture, the a-helix- predicting network uses the (i,i±4) energy term of the query sequence as the additional input, while the beta-predicting network uses >(i,i±4) energy term as the additional input.
During operation, the additional
input typically will span the range of scaled energy values
from -2.0 to 2.0. Because individual amino acids differ with respect to
side-chain length, composition, and hydrophobicity, the interaction
energy of an input sequence is pre-normalized by the average interaction
energy of the corresponding amino acids (See details in our publications).
Performance
Accuray of CSSP2 for predicting secondary structure at their native tertiary contexts
Traning set: 440,884 fragments of 7-residue length from SCOP20
Test set : 22,707 fragments extracted from 1,629
unique fold SCOP domains.
Single neural network : 74%
Dual neural network : 83%
(Find details in Yoon et al., Comp Biol & Chem, 2007)
Accuray for predicting aggregation-prone amino acid fragments
NetCSSP calculates P(helix), P(beta) and P(coil) for each residue of a given sequence
The amyloidogenic hidden beta propensity (HbP) is calculated using the form,
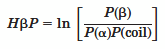
In this validation using ROC plots, the calculated "HbP" successfully priotizes aggregration-prone fragments over non-aggregates.
Test set 1 includes 104 fragments
Test set 2 includes 70 fragments
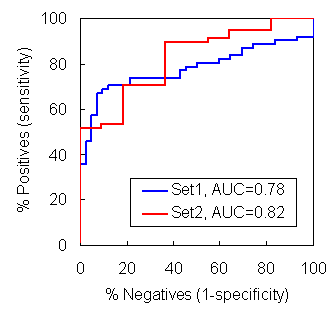
AUC (Area Under the Curve) represents the predictive power of the method
Test data sets were retrieved from Fernandez-Excamilla et al. (2004) Nat. Biotechnol., 22, 1302-1306
Pinpointing the core of amyloid fibril formation
Top figure: The CSSP profile of human IAPP4-34 sequence
Bottom figure: Summary of aggregation data of 28 consecutive overlapping decamers from IAPP (Raw data retrieved from Mazor et al. J. Mol Biol, 2002)
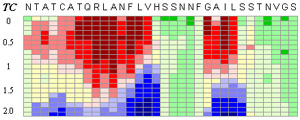
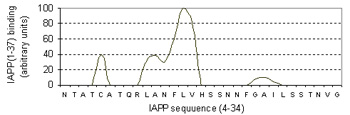
Energy-dependent fluctuation of beta-propensity (blue) in the entire CSSP profile is highly consistent with experimental data showing the core subsequence for amyloid fibril formation.
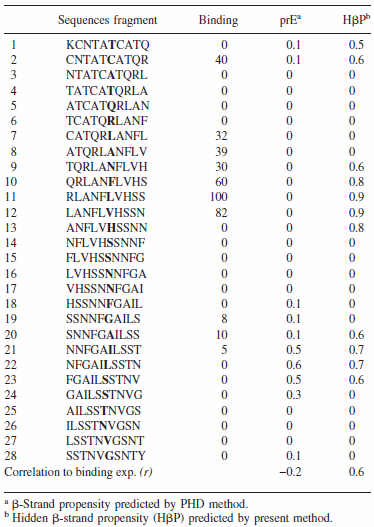
For 28 decamers, the calculated beta propensity is well correlated with the experimental relative aggragation ability, i.e., relative binding to amyloids.
(Find details in Yoon & Welsh, Protein Science, 2004 and Proteins, 2005)
Correlation between predicted "Hidden beta Propensity (HbP)" and experimental
aggregation rate of AcP mutants
Experimental data sets were retrieved from Chiti et al., Nat Struc Biol, 2002
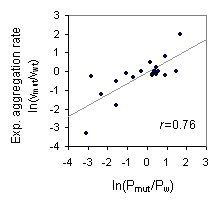
The Pmut and Pw represent mutant HbP and wild-type HbP respectively,
and they were calucalated using the form,
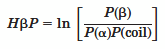 (Yoon and Welsh, 2005, Proteins)
(Yoon and Welsh, 2005, Proteins)
Comparison of CSSP-based beta propensity between amyloidogenic and non-amyloidogenic sub-sequences
Data from Yoon & Welsh, 2004, Protein Science
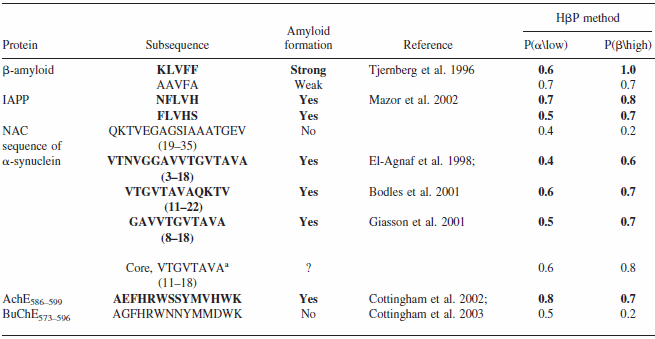
CSSP-based "hidden beta-propensity" in SCOP90 domains and known amyloidogenic proteins
Data from Yoon & Welsh, 2005, Poteins
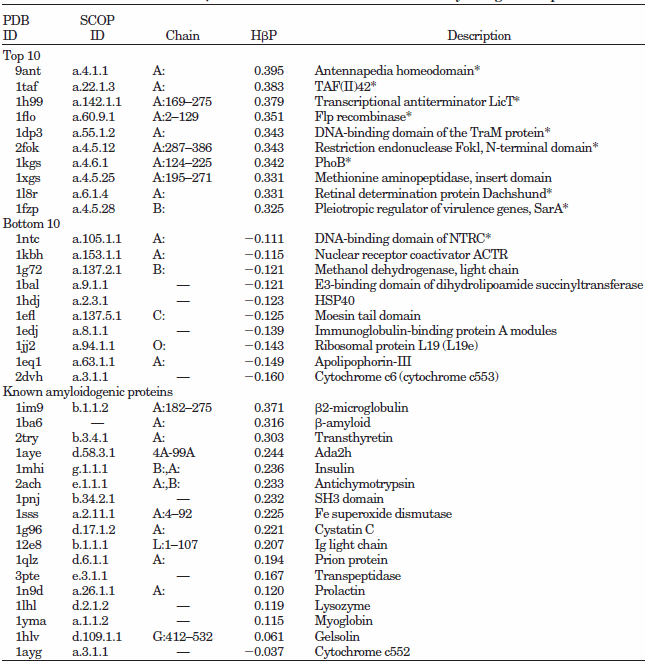
A total of 6,390 domains were retrieved from SCOP90v1.61